
Author: Doug Dollemore
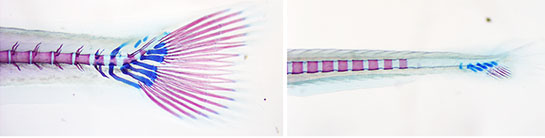
Fixing a design flaw in how CRISPRs are synthesized could help scientists more accurately determine the effects of genetic changes, such disabling tail development in zebrafish.
Sometimes close doesn’t quite cut it.
Take CRISPR, for instance. This remarkable gene-editing tool has revolutionized genetic research, allowing scientists to precisely rearrange DNA sequences and modify genes with ease. This emerging technology could enable researchers to repair genetic defects and develop new ways to prevent or even treat many diseases, including cancer. But sometimes it just doesn’t work as well as intended.
Why? According to David Grunwald, PhD, a professor of genetics at University of Utah Health, CRISPRs synthesized in the laboratory are just a wee bit too big.
More precisely, synthesized CRISPRs are commonly designed with one or two extra nucleotides at the end of their RNA, a form of genetic code. Normally, this doesn’t make much of a difference, but in some instances, it causes CRISPR to misalign with the DNA sequence it is trying to modify, producing unintended consequences. Removing those nucleotides from the CRISPRs, Grunwald’s group showed, significantly improves outcomes.
A report on this new CRISPR technology by Grunwald and his colleagues appears in Development Cell.
Grunwald says the finding could transform CRISPR research, allowing researchers to more accurately assess the effects of genetic modifications using a variety of animal models including fruit flies, zebra fish, and rodents. The discovery could also spur faster development of treatments for human genetic diseases.
In their study, Grunwald’s team found that redesigned CRISPRs cut 22 of 23 target genes with almost perfect efficiency. In contrast, only 4 of 10 CRISPRs made with the two extra nucleotides cut target genes with 100% efficiency. The other six worked modestly or poorly, leading to results that were difficult to interpret.
The discovery could spur faster development of treatments for human genetic diseases.
“Prior to this discovery, if I failed to get a result I wouldn’t know if the CRISPR failed or if the gene I was targeting wasn’t involved in the process I was interested in,” Grunwald says. “Now, with our new technique, it allows us to have a clear outcome nearly 100% of the time.”
Bacteria use this mechanism to identify potentially harmful viruses. Once the attacking virus is recognized, CRISPR guides an enzyme called Cas9 into place so it can shred viral DNA, rendering it harmless.
Scientists figured out how to replicate this process a few years ago, and soon afterward laboratories worldwide were using it to target unique DNA sequences within chromosomes to modify genes in animals and plants. As it does in bacteria, the synthesized CRISPR guides the Cas9 enzyme to a targeted section of DNA. There, Cas9 acts like a pair of genetic scissors, allowing scientists to disable (“knock out”), repair, or modify genes. In some cases, the CRISPR/Cas9 tool can even edit a single nucleotide, or base, within a DNA sequence.
To scientists, this gene-editing tool was almost miraculous, saving time and money in the lab. For Grunwald, who uses zebra fish in his genetic studies of osteoarthritis and osteoporosis, it meant that instead of tediously raising several generations of the fish to determine the effects of disabling a single gene—a process that can take months—he could use CRISPR/Cas9 to knock out one or more genes in a fish embryo and know what is happening within days.
However, Grunwald and others soon noticed that this genetic tool didn’t always work right. But instead of questioning how these synthesized CRISPR/Cas9 were made, most scientists accepted the tool would sometimes fail or produce inconclusive results.
“Their answer was to apply four different CRISPRs simultaneously to different parts of a gene and expect that the combined effects would eliminate the gene’s function,” Grunwald says. “So, they generally recognized there was a problem.”
Working with Integrated DNA Technologies (IDT), a company that creates CRISPR components, Grunwald and his colleagues sought to improve the consistency and efficacy of the mechanism. In experiments, they tested several different configurations of CRISPRs, each based on slightly different types of synthesized RNA molecules. Ultimately, they discovered the answer was a simple design oversight.
While the original method to synthesize CRISPRs was convenient and flexible, it also required that RNA molecules always begin with one or two guanine nucleotides. And that was its fatal flaw—it meant that laboratory-created CRISPR RNAs didn’t quite act like the protein-RNA complexes naturally produced by bacteria. As a result, the synthesized CRISPRs sometimes didn’t match up correctly with the DNA sequence they were targeting.
By removing the guanine bases at the beginning of the RNA, Grunwald’s team created more accurate CRISPRS that aligned perfectly with DNA sequences.
“The technique we developed is much cleaner, much more specific, and much more efficient at eliminating single gene activity,” Grunwald says. “Making CRISPR more precise and reducing the number of unintended effects is an incremental advance, but one that will have important impacts on medical research in the future.”