Jensen Lab Research

Jensen Lab Research
Hypoxia-inducible factor-1 in the metabolism and angiogenesis of intracranial tumors
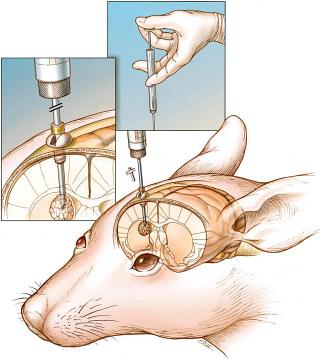
VEGF is important in the progression and angiogenesis of gliomas. Hypoxia-inducible factor-1 (HIF-1) is the major regulator of glycolysis and VEGF. Our current work looks at the role of HIF-1 and other hypoxia-regulated molecules in human brain tumors. Current work examines HIF-1 in malignant glioma development. We have made siRNA to inhibit the function of HIF-1 and are examining the effects of this methodology on brain tumor metabolism, vascularity and growth in a mouse model.
Calcium channel antagonists for potentiation of chemotherapy for meningiomas
Our initial work demonstrated the use of calcium channel antagonists for the growth inhibition of human meningiomas. This ongoing work involves using calcium channel antagonists to increase the efficacy of common chemotherapeutic drugs used for the treatment of meningiomas. This involves both cell culture experiments and an animal model of meningiomas first developed in our lab.
Post-treatment radiographic imaging changes (PTRIC)
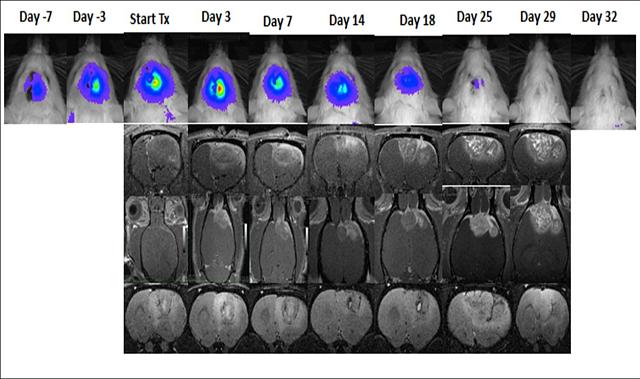
Glioblastoma multiforme (GBM), the most common and most malignant brain tumor in humans, has a dismal prognosis despite aggressive therapy. Initial diagnosis and measurement of response to treatment is usually determined by measurement of gadolinium-enhanced tumor volume with magnetic resonance imaging (MRI). Unfortunately, many GBM treatment modalities can cause changes in tumor gadolinium enhancement patterns that mimic tumor progression. These changes are usually found to improve or remain stable on follow-up MRI and have thus been termed “pseudoprogression”. The lack of a definitive imaging modality to distinguish PTRIC, including pseudoprogression and radiation necrosis, from true tumor progression presents a major unmet clinical need in the management of GBM patients. Furthermore, there is no clear understanding of the molecular, biochemical, and cellular mechanisms mediating PTRIC. We are developing a novel rodent model of PTRIC and applying new imaging modalities to distinguish PTRIC from true tumor progression.
Glioma stem cells and response to hypoxia
Recent advances in targeted therapy for cancer have had little influence on survival for patients diagnosed with Glioblastoma, and median survival time remains stagnant at 12-15 months post diagnosis. Genomic profiling of GBM indicates clear phenotypic subsets of these tumors, warranting the development of new models and targeted therapies. The mesenchymal phenotype in GBM is associated with poor patient prognosis and most recurrent GBM fall into this phenotype. Additionally, the cancer stem cell model applied to GBM explains the cellular heterogeneity of these tumors and their origin of growth. Glioma stem cells (GSCs) are thought to be responsible for propagation of the bulk tumor mass, resistance to chemotherapy and radiation treatment, and tumor recurrence and progression after surgical resection. We are investigating how regions of intratumoral hypoxia maintain the stem-like characteristics of GSCs and facilitate the shift to a mesenchymal phenotype, and how specific treatments targeting HIF-1 can effect tumor growth.
Other collaborative projects
The cancer stem cell research is done in collaboration with Dr. Howard Colman in the Dept. of Neurosurgery. This also involves cataloging surgical samples based on location and level of hypoxia to find relationships with other markers.
Our PTRIC research is a joint project with Drs. John Hoffman and Jeffrey Yap in Clinical Radiology
We are providing animal expertise in collaboration with Dr. Kuberan Balagurunathan’s research on custom designed heparin-based drug conjugates for use as drug delivery vehicles for cancer.
The Jensen Lab has worked with Drs. David Gaffney and Dennis Shrieve of the Radiation Oncology Department on a project measuring HIF-1 expression in cancer, and the radiobiology of meningiomas.
We have worked with Dr. Dennis Parker and Dr. Robert Roemer on a project for the “Optimization of Interactive Control of HIFU Therapy.”
RNAi
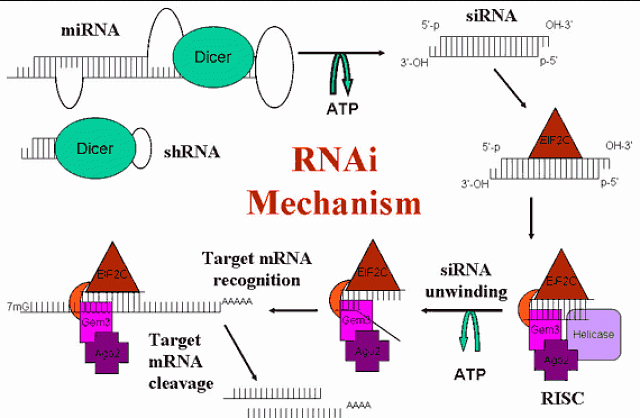
Figure 1: Diagram of the mechanism of RNAi. Naturally occurring microRNA (miRNA) and short hairpin RNA (shRNA) are processed by Dicer in the cytoplasm into short interfering RNAs (siRNA). These are recognized by EIF2C and other proteins of the RNA-induced silencing complex (RISC). The siRNA is unwound and used as a template for identifying a homologous mRNA, which is cleaved by an RNase H-like enzyme in RISC. The siRNA/RISC complex is then free for further mRNA cleavage.
Post-transcriptional gene silencing (PTGS) was first thought to be a novel mechanism limited to plants, but has been shown to be a highly conserved mechanism found also in worms, flies, and mammals. It appears to be an early defense mechanism against viruses and transposons, effectively silencing foreign and unwanted genes. The discovery of RNA interference (RNAi) in C. elegans using injected double strand RNA (dsRNA) launched the current wave of RNAi discoveries in mammals. Initially, these experiments failed due to the activation of the interferon response pathway by long dsRNA, which caused global inhibition of protein synthesis. This was overcome by Tom Tuschl's lab, which discovered that small RNAs of 19-23 bases, called short interfering RNA (siRNA), avoided this response. These siRNAs can be synthesized and introduced directly into cells (Figure 1). Unlike the extended silencing effect seen in worms and flies, the effect of siRNA in mammals is transitory, usually lasting up to 72 hours in cultured cells. However, the use of plasmids to express short hairpin RNA (shRNA), which mimics the recently discovered endogenous microRNA (miRNA), allows for extended gene silencing (figure 2).
RNAi is a potent tool for specific gene silencing that only requires the target gene sequence. Because of the sequence specificity, even different alleles of the same gene can be silenced. For a thorough review of RNAi, see Nature Insight, Vol. 431 pp. 338-378, 16 September, 2004.
We are using RNAi to inhibit HIF-1 and COX-II in vivo to determine the effects on glioma and meningioma tumor growth. We are also investigating delivery methods that will facilitate moving this potential therapy into a clinical setting.
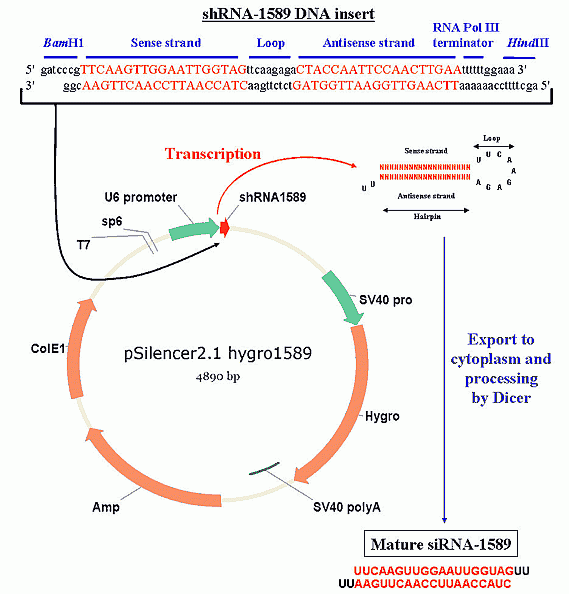
Figure 2: Diagram of RNAi mediated by shRNA production. First, a long dsDNA oligonucleotide is synthesized with the correct sequence (see methods for siRNA sequence selection) and cohesive ends, then inserted into a plasmid directly downstream of a RNA Pol III promoter (U6 or H1 are commonly used). The plasmid is amplified in bacteria, isolated and sequenced, then transfected into Your Favorite Cell line and stable clones are selected by antibiotic resistance (i.e., hygromycin). The U6 promoter constitutively expresses the shRNA insert, which folds into a hairpin structure and is exported from the nucleus. In the cytoplasm, Dicer recognizes the hairpin structure and processes it into a functional siRNA.